|
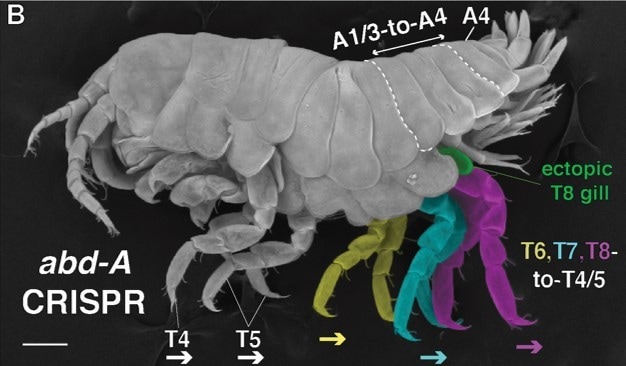
"Dr. Arnaud Martin details his research adapting the CRISPR-Cas9 gene editing system to crustaceans and butterflies, providing further evidence that supports previous findings surrounding the use of genetic tool kits found in all animals." Throughout evolutionary history, animals have used a variety of means to conquer new environments. From the smallest wasp to the great blue whale, animals have become masters at surviving just about anywhere on the planet. Some of this survival has been due to chance: a mutation here, a population bottleneck there. Though life is much cleverer still with yet greater schemes for allowing species to radiate into new realms. One such scheme is a simple set of genes shared by all known animal life on earth called Hox genes. What makes this collection of genetic material so clever is when and how it is used. Hox genes are expressed in variable combinations and locations during an animal’s developmental period, which eventually leads to the amazing variety of segmented body plans we see today. The number legs on a crab, the placement of antennae on a house fly, and even the arrangement of vertebrae in your spine are all determined by combinations of Hox genes. Developmental biologists have been studying Hox genes for quite some time and a great deal is known about them. New technology, however, must be adapted to previously studied systems and all scientific theory must be tested and retested constantly. In his recent talk at the University of Maryland, Dr. Arnaud Martin, an assistant professor at George Washington University, sought to do exactly that using the CRISPR-Cas9 system, a precise gene-editing tool.
Dr. Martin then turned his sights to a new animal that exploits a different evolutionary toolkit: the Heliconius butterfly. This genus of butterfly is interesting because the patterns on its wings mimic that of other local species, all of which warn predators that they are poisonous, including Heliconius. Species outside and within the Heliconius genus mimic each other’s wing patterns, despite being separated by many millions of years of evolution (Figure 2).
Today Dr. Martin is working on compiling his work, along with many others, into a genetic reference database called GePheBase www.gephebase.org. Specifically this database will contain evolutionarily relevant cases of how genes in a variety or organisms give rise to their observable traits, or phenotypes. The goal of which is to draw major evolutionary conclusions through a comparative approach. Currently GePheBase has over 1400 entries covering a variety of traits.
About the author: Anthony Nearman is a graduate student in Dr. Dennis vanEngelsdorp’s lab studying Honey bee genetics and their innate immune systems. He is currently working on discovering the cause and mechanism of nodule formation in the abdomen of Honey bees and variation of wing venation patterns that may arise from pathogen stress. References: Martin, Arnaud et al. "CRISPR/Cas9 mutagenesis reveals versatile roles of Hox genes in crustacean limb specification and evolution." Current Biology 26.1 (2016): 14-26. Current Biology. 10 Dec. 2015. Web. 6 Feb. 2017. Martin, Arnaud et al. "Wnt signaling underlies evolution and development of the butterfly wing pattern symmetry systems." Developmental Biology 395.2 (2014): 367-78. Science Direct. 6 Sept. 2014. Web. 6 Feb. 2017. Comments are closed.
|
Categories
All
Archives
June 2024
|
Department of Entomology
University of Maryland
4112 Plant Sciences Building
College Park, MD 20742-4454
USA
Telephone: 301.405.3911
Fax: 301.314.9290
University of Maryland
4112 Plant Sciences Building
College Park, MD 20742-4454
USA
Telephone: 301.405.3911
Fax: 301.314.9290


 RSS Feed
RSS Feed




