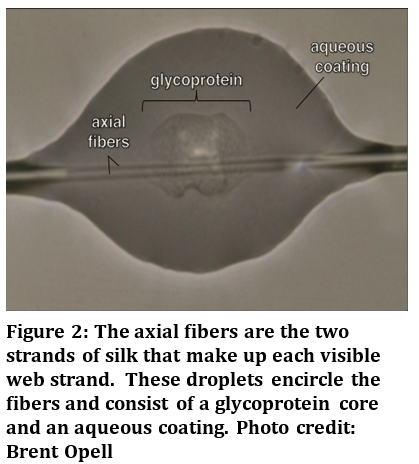
 UMD's Entomology Department professor Mike Raupp (pictured on the left) was spotlighted on the Today Show the morning of May 24, 2016 as he shared simple ways to combat mosquito breeding zones. Also discussed in the report is a comparison of natural mosquito repellents against those composed of synthetic chemicals. Be sure to check video here and leave us a comment of your thoughts on the growing concern of Zika virus carrying mosquitoes in United States. Many of us are familiar with spider webs in our gardens. You may have even heard that their silk, which makes up their webs, is among the strongest materials found on Earth. But we never think about how the web is actually covered in tiny sticky droplets (Figure 1). These droplets of glue have been the fascination of Dr. Sarah Stellwagen, a scientist at the Army Research Laboratory.
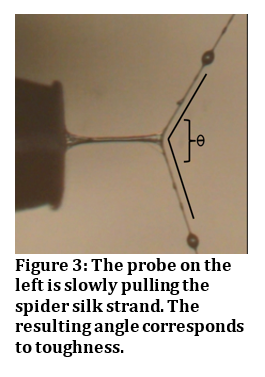
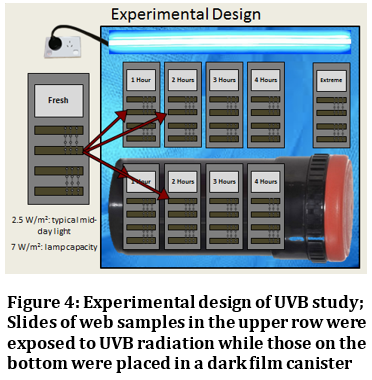
Dr. Stellwagen’s research on the various environmental factors that affect spider glycoprotein glue; namely temperature, humidity and UVB radiation, not only increases our understanding of spider biology but also holds promise for bioengineering efforts to mimic the amazing adhesive properties of spider glue.
References Stellwagen, S. D., Opell, B. D. and Short, K. G. (2014). Temperature mediates the effect of humidity on the viscoelasticity of glycoprotein glue within the droplets of an orb-weaving spider's prey capture threads. The Journal of Experimental Biology 217, 1563-1569. Stellwagen, S. D., Opell, B. D., & Clouse, M. E. (2015). The impact of UVB radiation on the glycoprotein glue of orb-weaving spider capture thread. The Journal of experimental biology, 218(17), 2675-2684. About the authors: Jessica Grant is a master’s student in Dr. Bill Lamp’s lab. Her work looks at the cold tolerance and phenology of kudzu bugs and how that impacts the range expansion and integrated pest management of the bug. For more information visit mdkudzubug.org Lisa Kuder is a PhD student in Dennis vanEngelsdorp’s Bee Lab. Working with the MD State Highway Administration she is evaluating sustainable pathways for supporting highway rights-of-way pollinator initiatives. Click here for details. Dr. Dawn Gundersen-Rindal is a researcher at the Invasive Insect Biocontrol and Behavior laboratory at the USDA ARS facility in Beltsville, Maryland. The goal of her team is to use biological, molecular, chemical and non-chemical techniques to develop novel, sustainable and cost-effective methods for the control of arthropod pests. They focus on invasive agricultural pests like the Brown Marmorated Stink Bug (Halyomorpha halys), medically important disease vectors like the Asian Tiger Mosquito (Aedes albopictus), and urban pests such as bed bugs (Climex lextularius). These pests in particular, have historically been difficult to control with some classes of traditional pesticides. Many of the pesticides available today are slow acting and persist in the environment for long periods of time. They are also indiscriminate, and will suppress beneficial arthropods, such as pollinators and natural enemies, and sometimes harm other organisms such as mammals, birds, and aquatic fauna. The advent of genetically modified crops allowed us to deliver toxins directly to the insects that were consuming the crop we wanted to protect. This directed approach protected the environment from broad applications of pesticides that were far more harmful to local ecosystems. However, they led to issues of resistance to the toxins used in GM plants and the potential of debris and the by-products from GM plants persisting the environment.
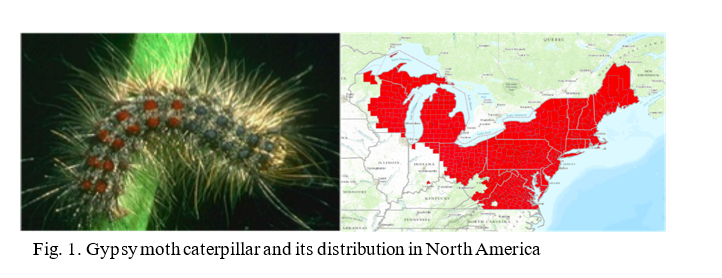
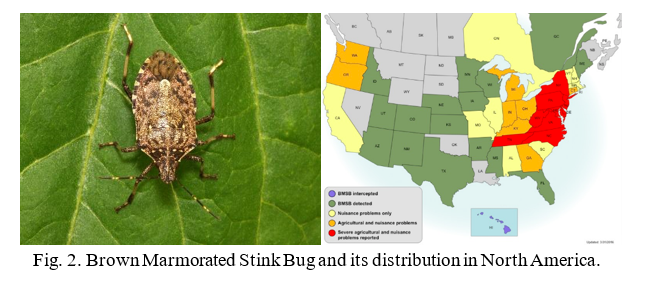
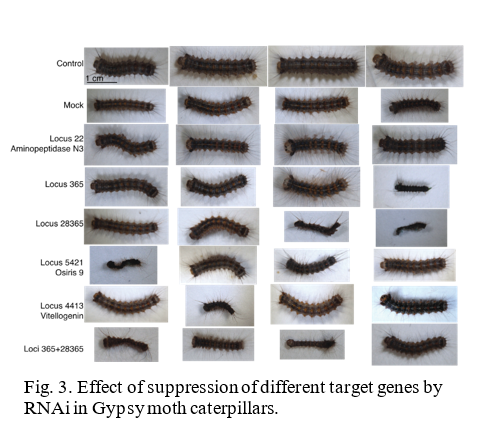
The lab is currently conducting RNAi research on two insects, the Gypsy moth (Fig.1) and the Brown Marmorated Stink Bug (BMSB) (Fig. 2). The European Gypsy moth (Lymantria dispar dispar) has been present in the US as an invasive pest since 1869, but has become a bigger threat with several recent accidental introductions and eradications of the Asian Gypsy Moth, (Lymantria dispar asiatica and other subspecies). The Asian gypsy moth is more destructive than its European counterpart due to the adult females having the ability to fly, allowing them to disperse much further, and the extensive number of trees and plants it defoliates. These traits are passed on to hybrid offspring of the two subspecies, further exacerbating this potential problem. BMSB, as mentioned previously, is a recent invasive that is an extremely damaging agricultural pest in the northeastern US. It has over 300 hosts and has caused billions of dollars in losses. It is also a nuisance pest as it overwinters within buildings. The USDA team focused on two requirements for developing RNAi control methods for these pests: 1) identifying suitable target genes and 2) finding an effective method for delivering dsRNA. The dsRNA is produced either through bacterial transformation (making bacteria produce mass quantities) or in vitro using PCR (using a machine to produce large quantities of the substance). The dsRNA is orally delivered through artificial diet in the lab. The minimum requirement for identifying target genes is a comparative transcriptome (a catalog of the messenger RNA expressed in a cell or group of cells), although a complete genome is preferred. The transcriptome used for the Gypsy Moth focuses on gene expression in response to midgut microbes. Preliminary trials did suppress some of the targeted gut genes in the Gypsy Moth, but this did not have a significant effect on development or mortality. Because RNAi can only suppress gene to a certain extent (maximum so far is 80%), it is necessary to identify genes important enough that partial silencing will cause severe damage. In the Gypsy moth, the most promising results came from two genes of unknown function, which led to a significant reduction in egg mass production. BMSB research is currently being conducted using two transcriptomes, and the whole genome is also being assembled. Although less dsRNA is required to kill BMSB with gene suppression than the Gypsy moth, delivering dsRNA is more difficult because it eats using a needle-like stylet, making an artificial diet or spray impractical. The team has developed a method to feed sufficient dsRNA to the BMSB for gene suppression, and is currently testing RNAi against different target genes in this species. These trials are currently in their initial phase and are expected to progress faster than the gypsy moth research due to the ease in causing mortality.
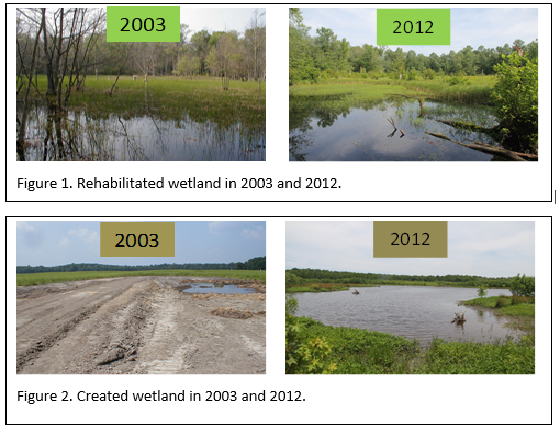
RNAi is an extremely promising pest management technique, but it is still in the preliminary stages of development. After a formulation is developed for field application, issues such as persistence in the environment and non-target impacts will need to be researched thoroughly before EPA approval can be granted. Public acceptance may also be a hurdle, especially as the first variants of these RNAi control methods may find themselves in transgenic crops. Although RNAi-based insecticides will not be commercially available for several years, their many positive attributes in comparison to conventional insecticides hold the promise of revolutionizing arthropod pest control. Further information on the transcriptomes used to develop RNAi techniques for both species can be found in the following publications: Sparks ME, Shelby KS, Kuhar D, Gundersen-Rindal DE. Transcriptome of the Invasive Brown Marmorated Stink Bug, Halyomorpha halys (Stål) (Heteroptera: Pentatomidae). Mittapalli O, ed. PLoS ONE. 2014;9(11):e111646. doi:10.1371/journal.pone.0111646. Sparks ME, Blackburn MB, Kuhar D, Gundersen-Rindal DE. Transcriptome of the Lymantria dispar (Gypsy Moth) Larval Midgut in Response to Infection by Bacillus thuringiensis. Terenius O, ed. PLoS ONE. 2013;8(5):e61190. doi:10.1371/journal.pone.0061190. Armando Rosario-Lebron: Is a graduate Student in the laboratory of Dr. Cerruti RR. Hooks at the University of Maryland that works on sustainable agriculture through cover crop management techniques. In the past he worked on genetic disorders in the eye in the laboratory of Dr. Jaqueline Tanaka at Temple University. Aditi Dubey is a PhD student in the Hamby Lab, studying non-target effects and sustainable use of neonicotinoid seed treatments.  Congratulations to Dr. Elanor Spadafora (Lamp Lab), the UMD BISI Graduate Program's newest PhD graduate, for successfully defending her doctoral dissertation: "Analysis of macroinvertebrate communities In seasonal wetlands through time, space, and species traits" Check out a recent UMD Entomology Colloquium Blog post about her research here! Best of luck to to Dr. Spadafora as she continues her career! If you take a drive along the Eastern Shore of Maryland, you will likely pass many wetlands. Called Delmarva bays, these wetlands are seasonal, meaning that they dry out every summer. Consequently, fish cannot survive in most of these wetlands, and the food webs are mainly composed of aquatic invertebrates. Delmarva bays have become the focus of recent restoration efforts under organizations such as the Nature Conservancy. Over the years, many of these wetlands have been impaired or destroyed by agricultural activity. This is problematic, as Delmarva bays perform many important ecosystem services, such as retaining nutrients that would otherwise enter the Chesapeake Bay. A central goal of restoration efforts is to reestablish these services. As PhD candidate, Ellie Spadafora of the Lamp Lab learned, however, determining the success of wetland restoration can depend a lot on when and how you measure it. Spadafora first wanted to examine the community level changes between three types of wetlands: natural, rehabilitated, and created. The rehabilitated and created wetlands differed in that the rehabilitated wetland had once been a wetland in the past while the created wetland was constructed in a forested depression. The created and rehabilitated wetlands were restored and built, respectively, in 2003, and monitoring of invertebrate populations began soon after. How much had they changed, and begun to resemble the historic wetland, from 2003 until the start of her research in 2012? Visually, the restored wetlands looked much healthier after 9 years, as seen in the comparison in figures 1 and 2. Do these visual changes mean anything for the biological composition of the wetlands? Spadafora compared the aquatic invertebrates sampled in the wetlands over time and found some encouraging results. The most abundant invertebrate in the natural wetland across all years was a freshwater isopod, Asellus. The created wetland, in contrast, had non-biting midge larvae (Family Chironomidae) in abundance every year. The rehabilitated wetland was the most interesting. Although in its earlier years the invertebrate community was dominated by midge larvae, by 2012 Asellus accounted for the majority of the specimens. This may suggest that if the goal of a wetland restoration project is to achieve similar characteristics to natural wetlands, rehabilitating former wetlands may be a better plan than creating new ones. However, her results did indicate that success cannot necessarily be determined immediately following restoration. Longer term monitoring may be needed to see improvements. So what was different about the rehabilitated and created wetlands? Spadafora hypothesized it may have something to do with a moss. Naturally occurring sphagnum moss (figure 3) may be most recognizable as a base for floral arrangements, but it also serves as an important ecosystem engineer by providing habitat structure for aquatic invertebrates, such as isopods and predaceous diving beetles (Family Dytiscidae, figure 4). Sphagnum also appeared in the rehabilitated wetland in 2009, which is the time period where isopods became dominant in the invertebrate community. To test her theory that sphagnum influences invertebrate communities, she expanded her study to include twenty Delmarva Bays (ten with sphagnum, ten without) and shifted her taxonomic focus to Dytiscid beetles. Although not as abundant as the isopods, these Dytiscid beetles are crucial for Delmarva bay ecosystems because, in the absence of fish, they are one of the top predators. If sphagnum mosses were influencing beetle diversity, she expected to see more species of beetles in the sphagnum wetlands. Indeed, beetle diversity was higher in the moss-containing wetlands, and interestingly some species were only found in either the sphagnum or non-sphagnum sites. Spadafora’s research provides us with some excellent takeaways regarding wetland restoration. In addition to making the rehabilitated wetland look more like a genuine Delmarva bay, the sphagnum may also allow it to support a larger array of aquatic insects and maintain diversity of a disappearing habitat type.
For more information, check out Ellie Spadafora’s recent publication in Restoration Ecology. About the author: Becca Wilson is a PhD candidate in the Lamp Lab. She studies the spatial distribution and societal impacts of nuisance black flies in western Maryland. Congratulations to Entomology’s Dennis vanEngelsdorp, Nathalie Steinhauer, Karen Rennich and their colleagues with the Bee Informed Partnership, whose latest annual survey results on honey bee colony losses are released today. Results of the survey suggest that beekeepers across the United States lost more than 44 percent of their honey bee colonies during the year spanning April 2015 to April 2016. Rates of both winter loss and summer loss—and consequently, total annual losses—worsened compared with last year. This marks the second consecutive survey year that summer loss rates rivaled winter loss rates.
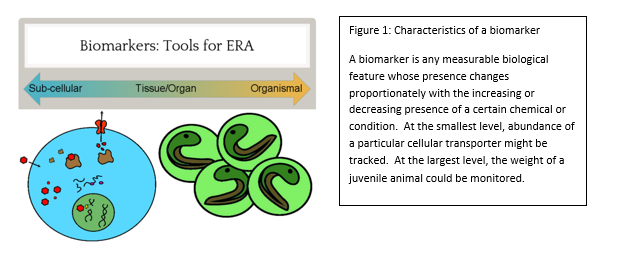
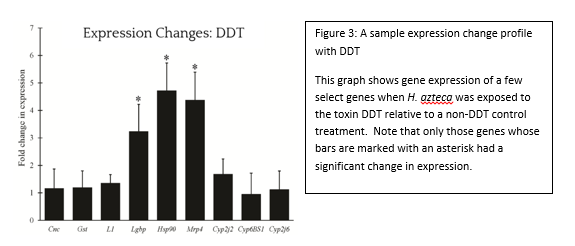
Check out the news release posted on the CMNS website : https://cmns.umd.edu/news-events/features/3539 There have been numerous media inquiries. The story Minnesota Public Radio can be found here: http://www.mprnews.org/story/2016/05/10/bee-colony-losses-up-study-says From simple metal ions to complex endotoxins of bacterial origin, poisons have the potential to wreak havoc at every biological level from cell to ecosystem. This potential did not go unnoticed by cunning humans, some of whom educated themselves in this matter to the point of becoming assassins by way of poison. However, the pernicious effects of toxic chemicals on the environment, like the impact of DDT on bird eggshells, were eventually realized. This spurred the study of chemicals in the environment to prevent or fix ecological disasters and ultimately gave rise to agencies such as the EPA, and the subfield of ecotoxicology as a whole. In order to study the effects of a chemical on the environment, including the organisms that inhabit it, an Ecological Risk Assessment (ERA) is undertaken. One of the tools of an ERA is the use of biomarkers, which are measurable changes in certain aspects of a cell, tissue, organ, or organism that can accurately and consistently indicate the presence and/or activity of a particular chemical pollutant (see figure 1). Although we disproportionately care more about large animals such as deer, bears, and fish, tiny organisms are the preferred indicators, because they are plentiful and easy to work with. In aquatic environments, we have traditionally exploited the water flea, Daphnia, and midge larvae of the family Chironomidae. However, the amphipod Hyalella azteca has been gaining traction due to its unique ecological niche. The fact that this species dwells at the soil-water interface makes it an ideal candidate to represent the toxicological conditions at this particular location. Macroscopic changes such as lifespan and reproduction have been the gold standard of ecotoxicology because they directly relate to adverse effects on populations, the real concern during ERA. However, the molecular revolution has ushered in many advances and we have progressed to the point of using expressed genes as biomarkers. Genetic biomarkers, as compared to gross morphological and ecological ones, have a much lower threshold of detection for toxins in the environment and therefore can be utilized in a novel and desirable approach. The particular use of these indicators in H. azteca has been the main focus of ecotoxicologist Dr. Ryan Gott, of the University of Maryland’s Department of Entomology . His multi-year investigation, which was completed in the lab of Dr. William Lampwas structured to elucidate several genetic biomarkers in this crustacean: which gene products can serve as faithful biomarkers, how to properly control the quality of experiments, and what spectrum of chemicals can biomarkers in this organism reliably report? The first stages of his work required generating a slew of data which describe genes that are normally expressed in a controlled environment, a body of biological information known as the transcriptome. These data, which are generated through a series of sophisticated molecular and computational techniques, enabled Gott to survey the transcriptome and assign different functions to all recognized genes. The next step in this scheme was to determine which H. azteca genes are most suitable to be monitored, as well as which can serve as controls. The genes chosen for monitoring were largely based off of those in Daphnia which encode proteins that respond to chemical stressors. Now, the bulk of the research could begin. Gott chose several chemicals that were classified as either metals (cadmium and copper) or organic pesticides (permethrin, DDT and imidacloprid). The first two were chosen due to the fact that man-made structures like refineries and power plants can concentrate them at levels much higher than those at a typical background level. The latter three were selected because insecticides are very widely used and can easily be distributed in bodies of water through pesticide drift. However, both toxin types can lead to a litany of cellular and physiological problems. After completing his experiments, Dr. Gott found some very interesting results during the analysis phase. Not only were there clearly observable changes in gene expression after chemical exposure, but several of the genes that responded to the metals also responded to the insecticides (see figure 3). Those genes whose expression was altered include cnc, which codes for a protein responsible for activating other proteins which handle stressors, Hsp90, a protein chaperone, and Mrp4, a transporter protein that moves molecules across the cell’s membrane. Dr. Gott’s highly practical research has convincingly shown that one can monitor changes in gene expression in H. azteca to make inferences about how organisms are being affected by potential toxins in the environment. The ability to track toxin dynamics that are too minute to be observed through conventional aspects such as the body size of an organism is a powerful advantage to this still-developing molecular approach. One of the major obstacles Dr. Gott and similar researchers face, though, is the ability to accurately link these small changes in the organism with the broad and very obvious changes in populations and ecosystems we ultimately care about. One solution that is likely to facilitate progress here is nailing down the relationships between cell, organism, and ecosystem first and then run experiments to see how these relationships are perturbed. Regardless of any barriers at the moment, this field of research is sure to greatly assist us in preventing, and hopefully ending, the catastrophes that toxins continue to create.
About the author: Justin Rosenthal is a 4th year PhD. Candidate studying under Dr. Jian Wang, and is also in the Entomology Department at the UMD. He is using the Drosophila model system to uncover important signaling pathways involved in nervous system development, specifically at the level of the individual neuron.  In two recent reports, NBC News reached out to UMD Entomology's Dr. Mike Raupp regarding how to safeguard yourself and others against mosquitoes this season as the threat of the mosquito-borne Zika virus may evidently pose a threat to humans in the US. Strategies include 1) wearing mosquito-repellent clothing, and 2) removing even the smallest possible breeding sites from around your home. Click here and here for the both ABC News online reports.  Dr. Margaret Palmer, UMD Entomology Professor and Director of the National Socio-environmental Synthesis Center (SESYNC), along with her colleague J.B. Ruhl, was awarded for the 2016 Sustainability Award from the Ecological Society of America (ESA) for their 2015 paper published in Frontiers in Ecology and the Environment entitled "Aligning restoration science and the law to sustain ecological infrastructure for the future." Click here to see the full citation. According to ESA, "The Sustainability Science Award is given to the authors of a scholarly work that makes the greatest contribution to the emerging science of ecosystem and regional sustainability through the integration of ecological and social sciences." Dr. Palmer and her colleagues also recently published a paper in the April 2016 edition of Current Opinion in Environmental Sustainability entitled "Socio-Environmental Systems (SES) Research: what have we learned and how can we use this information in future research programs". Click here to see the full citation. Congratulations Dr. Palmer!  The red flour beetle consuming stored grain The red flour beetle consuming stored grain Today, the red flour beetle, Tribolium castaneum, is not only a ravenous stored grain-eating pest, it is also a model organism for different study areas. Dr. Sue Brown is a University Distinguished professor of Kansas State University focusing on Genomics and Bioinformatics of Tribolium castaneum as a model system. She spent the beginning of her scientific career developing this beetle as a model for studying evolution of gene regulatory networks. She woke up every morning curious about the development and evolution of these charismatic little beetles, but she spent her evenings frustrated by her sometimes-functional degenerate primers, which she used to amplify and study Tribolium genes. As the name implies, degenerate primers are not ideal: these primers are designed from the genes of another organisms that are hopefully similar enough to be used on a gene from an organism of choice. However, without a genome sequence for Tribolium, this was as good as Brown could do. Tired of jumping over this hurdle, she led the effort to sequence Tribolium castaneum, the first beetle and the fourth insect ever sequenced [1]. Brown admits that “people come to genome projects from different points of view, but a lot of us are coming to get at the genes and the regions with the genes we are interested in so we can get back to the basic biology which got us interested in the first place.” She also admits, “DNA is very seductive,” which is why she has lingered trying to improve the quality of red flour beetle (and many other) genomes in lieu of jumping back into basic biology.  A Desktop BioNano sequencing machine. A Desktop BioNano sequencing machine. At the UMD Entomology department colloquium, Brown outlined how BioNano optical maps have improved the assembly of her beetle genome [2]. Before we discuss how the genome is improved, it is important to know why genomes need improvement in the first place. You can think of the genome as a book containing the information required to make a particular organism. In this metaphor, the genes are sentences of the book: they contain useful information, but are only a part of the larger story of the organism. The majority of genomes that have been sequenced are called “draft genomes.” They are called drafts because they are incomplete, and the most common reason they are incomplete is because they usually contain many gaps resulting in a genome composed of many fragments of an entire genome. A draft genome with eight chromosomes, or parts, might be composed of over 1000 fragments. Now, imagine trying to read a book composed of 1000 shuffled segments with sections missing. Perhaps you will find a quote or sentence you are particularly interested in, but the story certainly will not be the same. This is the issue facing scientists today: with most of our sequenced genomes very fragmented, we can find some genes of interest, but the story of how the genome is connected and how they differ among organisms is lost in the gaps. Dr. Sue Brown thinks that BioNano optical maps will contribute to the solution. Brown says, “in my lab everything old is new” because she is using a very old idea with a modern twist to stitch the fragments we have of genomes together. The old idea is called DNA fingerprinting, and the modern twist is fluorescent labeling of DNA using restriction enzymes that have been modified to nick the DNA where she wants to mark it. First, Brown strikes a delicate balance trying to obtain clean and ultra-long DNA for her optical maps: “to get it ultra-long, you want to manipulate it as little as possible; to get it ultra-clean, you want to manipulate it as much as possible.” She can then use specific enzymes to find, nick and replace small sections of the DNA with fluorescent nucleotides across the entire genome. Imagine highlighting a single word throughout an entire book. What you will produce is a barcode: page fifteen might have 3 highlights, page sixteen might have 1, page seventeen might have none, page eighteen might have 2, and so on. Having this barcode allows you to line up and extend long sections of your book to completion. Once Brown has marked the long segments of DNA, she can read each long barcode by running the DNA through a small imaging chip and line up the patterns. With enough long sequences, she can theoretically line up the entire genome in barcode. By comparing these barcodes with one inferred from sequence data, Brown can validate, but more importantly, stitch together the sequenced fragments of genomes. After choosing the optimum length for her barcodes and for the fragments from other technologies, she increased the average length of the Tribolium genome fragments from 1Mb to 4Mb. She is now working to apply this technology to many, often ignored, insects and organisms with low medical importance like cats and strawberries. Brown hopes to apply BioNano optical mapping to improve genomic studies of many different organisms, and she is constantly trying to optimize and develop techniques to advance genomic study of these organisms. Brown has come a long way studying red flour beetle segmentation, through the A’s, T’s, C’s, and G’s of DNA, to the 1’s and 0’s of barcoding. If her history ushering a humble red beetle from an emergent model organism into a bona fide model is any indication, Brown will not be satisfied with her development of the emerging BioNano technology until it becomes a standard practice in genomics. Reference 1. Richards, S., Gibbs, R. A., Weinstock, G. M., Brown, S. J., Denell, R., Beeman, R. W., ... & Klingler, M. (2008). The genome of the model beetle and pest Tribolium castaneum. Nature, 452(7190), 949-955. 2. Shelton, J. M., Coleman, M. C., Herndon, N., Lu, N., Lam, E. T., Anantharaman, T., ... & Brown, S. J. (2015). Tools and pipelines for BioNano data: molecule assembly pipeline and FASTA super scaffolding tool. BMC genomics, 16(1), 1. About the authors: Brian Lovett is a PhD student studying mycology and genetics in agricultural and vector biology systems. He is currently working on projects analyzing mycorrhizal interactions in agricultural systems, the transcriptomics of malaria vector mosquitoes, and the genomes of entomopathogenic fungi. Mengyao Chen is a Master’s student in Dr. Leslie Pick’s Lab. Her research focuses on segmentation genes in Brown Marmorated Stink Bug (BMSB, Halyomorpha halys). Her current work is looking for orthologs of pair-rule genes in BMSB, and studying their expression and functions using in situ hybridization and RNAi.  Click the link below to listen to Chloe Garfinkel - undergraduate researcher in the Lamp Lab - describe a hellgrammite and a crayfish to visitors at the "Discover a Swamp" exhibit at the 2016 Maryland Day! Watch the complete video here! |
Categories
All
Archives
June 2024
|
Department of Entomology
University of Maryland
4112 Plant Sciences Building
College Park, MD 20742-4454
USA
Telephone: 301.405.3911
Fax: 301.314.9290
University of Maryland
4112 Plant Sciences Building
College Park, MD 20742-4454
USA
Telephone: 301.405.3911
Fax: 301.314.9290



 RSS Feed
RSS Feed




