|
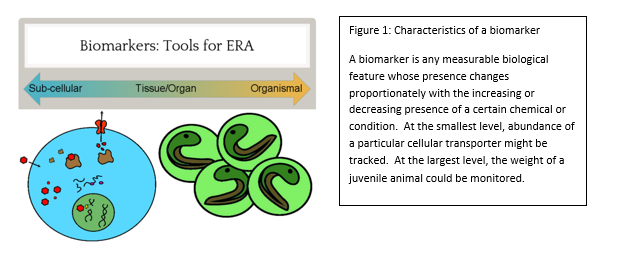
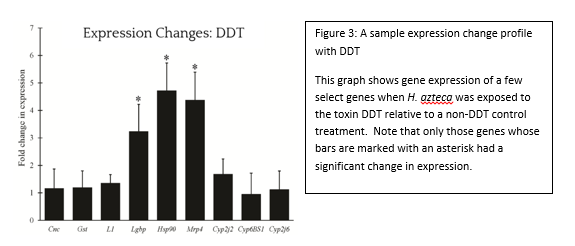
From simple metal ions to complex endotoxins of bacterial origin, poisons have the potential to wreak havoc at every biological level from cell to ecosystem. This potential did not go unnoticed by cunning humans, some of whom educated themselves in this matter to the point of becoming assassins by way of poison. However, the pernicious effects of toxic chemicals on the environment, like the impact of DDT on bird eggshells, were eventually realized. This spurred the study of chemicals in the environment to prevent or fix ecological disasters and ultimately gave rise to agencies such as the EPA, and the subfield of ecotoxicology as a whole. In order to study the effects of a chemical on the environment, including the organisms that inhabit it, an Ecological Risk Assessment (ERA) is undertaken. One of the tools of an ERA is the use of biomarkers, which are measurable changes in certain aspects of a cell, tissue, organ, or organism that can accurately and consistently indicate the presence and/or activity of a particular chemical pollutant (see figure 1). Although we disproportionately care more about large animals such as deer, bears, and fish, tiny organisms are the preferred indicators, because they are plentiful and easy to work with. In aquatic environments, we have traditionally exploited the water flea, Daphnia, and midge larvae of the family Chironomidae. However, the amphipod Hyalella azteca has been gaining traction due to its unique ecological niche. The fact that this species dwells at the soil-water interface makes it an ideal candidate to represent the toxicological conditions at this particular location. Macroscopic changes such as lifespan and reproduction have been the gold standard of ecotoxicology because they directly relate to adverse effects on populations, the real concern during ERA. However, the molecular revolution has ushered in many advances and we have progressed to the point of using expressed genes as biomarkers. Genetic biomarkers, as compared to gross morphological and ecological ones, have a much lower threshold of detection for toxins in the environment and therefore can be utilized in a novel and desirable approach. The particular use of these indicators in H. azteca has been the main focus of ecotoxicologist Dr. Ryan Gott, of the University of Maryland’s Department of Entomology . His multi-year investigation, which was completed in the lab of Dr. William Lampwas structured to elucidate several genetic biomarkers in this crustacean: which gene products can serve as faithful biomarkers, how to properly control the quality of experiments, and what spectrum of chemicals can biomarkers in this organism reliably report? The first stages of his work required generating a slew of data which describe genes that are normally expressed in a controlled environment, a body of biological information known as the transcriptome. These data, which are generated through a series of sophisticated molecular and computational techniques, enabled Gott to survey the transcriptome and assign different functions to all recognized genes. The next step in this scheme was to determine which H. azteca genes are most suitable to be monitored, as well as which can serve as controls. The genes chosen for monitoring were largely based off of those in Daphnia which encode proteins that respond to chemical stressors. Now, the bulk of the research could begin. Gott chose several chemicals that were classified as either metals (cadmium and copper) or organic pesticides (permethrin, DDT and imidacloprid). The first two were chosen due to the fact that man-made structures like refineries and power plants can concentrate them at levels much higher than those at a typical background level. The latter three were selected because insecticides are very widely used and can easily be distributed in bodies of water through pesticide drift. However, both toxin types can lead to a litany of cellular and physiological problems. After completing his experiments, Dr. Gott found some very interesting results during the analysis phase. Not only were there clearly observable changes in gene expression after chemical exposure, but several of the genes that responded to the metals also responded to the insecticides (see figure 3). Those genes whose expression was altered include cnc, which codes for a protein responsible for activating other proteins which handle stressors, Hsp90, a protein chaperone, and Mrp4, a transporter protein that moves molecules across the cell’s membrane. Dr. Gott’s highly practical research has convincingly shown that one can monitor changes in gene expression in H. azteca to make inferences about how organisms are being affected by potential toxins in the environment. The ability to track toxin dynamics that are too minute to be observed through conventional aspects such as the body size of an organism is a powerful advantage to this still-developing molecular approach. One of the major obstacles Dr. Gott and similar researchers face, though, is the ability to accurately link these small changes in the organism with the broad and very obvious changes in populations and ecosystems we ultimately care about. One solution that is likely to facilitate progress here is nailing down the relationships between cell, organism, and ecosystem first and then run experiments to see how these relationships are perturbed. Regardless of any barriers at the moment, this field of research is sure to greatly assist us in preventing, and hopefully ending, the catastrophes that toxins continue to create.
About the author: Justin Rosenthal is a 4th year PhD. Candidate studying under Dr. Jian Wang, and is also in the Entomology Department at the UMD. He is using the Drosophila model system to uncover important signaling pathways involved in nervous system development, specifically at the level of the individual neuron. Comments are closed.
|
Categories
All
Archives
June 2024
|
Department of Entomology
University of Maryland
4112 Plant Sciences Building
College Park, MD 20742-4454
USA
Telephone: 301.405.3911
Fax: 301.314.9290
University of Maryland
4112 Plant Sciences Building
College Park, MD 20742-4454
USA
Telephone: 301.405.3911
Fax: 301.314.9290

 RSS Feed
RSS Feed




