Phylogenetics and Bioinformatics Pair to Unravel Trichopteran Evolutionary Mysteries  Fig. 1: A drawer from the Smithsonian Institution’s Trichoptera collection (Frandsen 2015) Fig. 1: A drawer from the Smithsonian Institution’s Trichoptera collection (Frandsen 2015) Trichoptera, or caddisflies, comprise the most diverse strictly aquatic insect order. The desire to understand the cryptic evolutionary background of such incredible diversity has inspired an eager phylogeneticist, Dr. Paul Frandsen, to research Trichoptera. As Dr. Frandsen began his talk about Trichoptera, it became clear that he was fascinated by their wide-ranging behaviors and appearances (Fig. 1). Listening to him talk, I began to share his fascination. Trichoptera has three suborders: Integripalpia, Annulipalpia, and Spicipalpia. Integripalpia create mobile homes out of passing junk such as twigs, rocks, and tubes. Some Integripalpia even develop a fondness for bling and make a home out of jewels.  Fig. 2: Trichoptera larvae making jewelry (Matus 2012) Fig. 2: Trichoptera larvae making jewelry (Matus 2012) These caddisfly larvae are great jewelry-makers (Fig. 2)! Annulipalpia also known as the “fixed-retreat makers” are unique for their ability to spin elaborate capture nets, filters, and funnels out of silk. Dr. Frandsen described some of their living quarters as appearing “like twigs sticking up in sandy soil”. Larvae of Spicipalpia range from making cases to being free-living, but most make cases as pupae. In terms of feeding, some Trichoptera are filter-feeders while others are predators. Dr. Frandsen is particularly interested in trying to discern the evolutionary divergence of families of Spicipalpia, a suborder that is still taxonomically unclear. The families of Spicipalpia are Rhyacophilidae, Hydrobiosidae, Glossosomatidae, Hydroptilidae, and Ptilocolepidae. 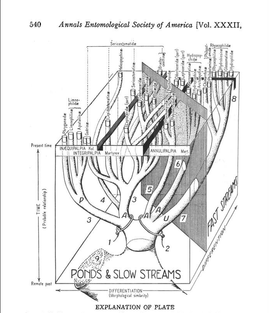 Fig. 3: Diagram showing the evolutionary differentiation of the suborders of Trichoptera from ponds to faster streams (Milne and Milne 1939) Fig. 3: Diagram showing the evolutionary differentiation of the suborders of Trichoptera from ponds to faster streams (Milne and Milne 1939) The process of resolving the “story” of how these families evolved currently draws from disciplines and techniques more aligned with the cloud computing technology of tech giants than traditional scrutiny of insect samples from museums. However, the first record of attempting to understand Trichoptera evolution appears in a paper by Milne and Milne in 1939, in which the authors proposed an evolutionary history where Trichoptera moved from ponds to slow moving streams to fast moving streams (Fig. 3). The authors of that paper considered two situations where Trichoptera anatomy “fit” the story. In their hypothesis the Rhyacophilidae are considered a more recently diverged family of Trichoptera. Currently, entomologists still have not completely resolved the evolutionary history of Trichoptera, but alternative evolutionary trajectories that suggest Rhyacophilidae are actually more ancestral have been supported by recent studies (Malm et al. 2013). 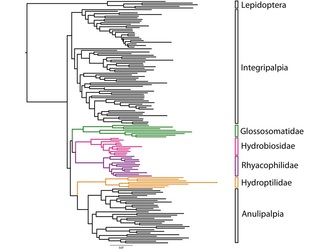 Fig. 4: Phylogenetic tree of Trichoptera families created using the k-means algorithm for individual DNA patterns in genes (Frandsen et al. 2015). Fig. 4: Phylogenetic tree of Trichoptera families created using the k-means algorithm for individual DNA patterns in genes (Frandsen et al. 2015). In an effort to resolve disagreements about how these organisms are related to each other, Dr. Frandsen has decided to tackle the problem using modern biotechnological techniques. DNA was sequenced en mass among many species. The data that resulted from this mass genetic analysis of these organisms was enormous. When different analysis and modeling techniques were used to map the relationships between Trichoptera, a puzzling inconsistency was found between data sets. Each technique seemed to be telling a different story that did not match what was previously known about Trichoptera biology and appearance. Dr. Frandsen then decided to collect even more samples, this time using DNA protein-coding regions called exons. This technique is unique as it only selects areas that are part of the final mRNA (after editing) used to transcribe proteins. This again yielded another enormous dataset that required super-computing effort to decipher. In order to do this, Dr. Frandsen developed a new model selection algorithm (Frandsen et al. 2015). Essentially, he co-opted the k-means clustering algorithm used by technology companies to group “people” or groups of people by the trends those people are following. For Dr. Frandsen, the “trend” computed would be Trichopteran genes and instead of people, individual DNA patterns. For example, rather than grouping people who visit shoe websites together, the mathematical algorithm was used to determine which DNA sites have the most similar evolutionary patterns and thus should be modeled together. After analyzing the information generated from the algorithm, a phylogenetic tree was made to show the evolutionary relationships of Trichoptera taxa (Fig. 4). Dr. Frandsen was then able to address some of the ambiguity of the Spicipalpia families. Dr. Fransen’s phylogenetic tree (Figure 4) shows that Glossomatidae are a basal family. Hydrobiosidae and Rhyacophiladae diverged later. The Hydroptilidae diverged separately as an independent clade, suggesting that the Spicipalpia are polyphyletic. Currently, Dr. Frandsen is working on analyzing the evolutionary relationships between Trichopteran transcriptomes (the set of all RNA molecules transcribed in a cell or population of cells), and how Ptilocolepidae, an enigmatic Spicipalpian family, can be incorporated into the phylogenetic tree (Frandsen 2015). Results from this analysis provide a new picture of how caddisfly families relate to each other and reveals how big data may help answer questions on other cryptic species and overall evolutionary history of life on planet earth. Despite his extensive undertaking, Dr. Frandsen believes that there is still much more work that needs to be done. There is ongoing research to discover better ways to create models, develop algorithms that are faster and more accurate, and create hardware that can process much more data. The field of bioinformatics is exciting and still quite young, so the potential for entomological applications seems very promising. References: Frandsen et al. (2015), Automatic selection of partitioning schemes for phylogenetic analyses using iterative k-means clustering of site rates. BMC Evolutionary Biology, 15:13 Frandsen, Paul (2015), Phylogenetic model selection and data filtering on large data sets: a case study with the caddisflies (Insecta: Trichoptera). University of Maryland Colloquium. Malm, T., Johanson, K. A. and Wahlberg, N. (2013), The evolutionary history of Trichoptera (Insecta): A case of successful adaptation to life in freshwater. Systematic Entomology, 38: 459–473. doi: 10.1111/syen.12016 Matus, Morgana (2012), Artists Enlist Caddisfly Larvae to “Build” Jewelry From Gold, Gemstones. Ecouterre Milne, J. and M.J. Milne (1939), Evolutionary trends in caddis worm case construction. Annals of the Entomological Society of America, 32(3): 540, Fig. 1 About the Authors:
Hanna Kahl and Armando Rosario-Lebronare are graduate students at the University of Maryland in the Entomology Department. Rahul Raiker is an undergraduate student at the University of Maryland in the Molecular Biology Department. Comments are closed.
|
Categories
All
Archives
June 2024
|
Department of Entomology
University of Maryland
4112 Plant Sciences Building
College Park, MD 20742-4454
USA
Telephone: 301.405.3911
Fax: 301.314.9290
University of Maryland
4112 Plant Sciences Building
College Park, MD 20742-4454
USA
Telephone: 301.405.3911
Fax: 301.314.9290

 RSS Feed
RSS Feed




