|
Written by: Elizabeth Brandt, Mintong Nan, Anna Noreuil, Katie Reding How it is possible to maintain a segmented body plan after loss of a key developmental gene? Dr. Alys Jarvela, a biochemist, geneticist, and postdoctoral scholar in the Pick Lab at University of Maryland, presented her research to address this precise question. Dr. Jarvela first spoke about the body plan shared by all insects featuring a head, thorax, and abdomen, each of these regions being partitioned into smaller segmental units. The genetic mechanisms underlying Drosophila segmentation is well-studied. An egg isn’t just uniform throughout; there are specific proteins and mRNAs that are localized to different parts of the egg. As development proceeds, the embryo is broken up into more defined regions and eventually each part of the embryo is specified to make a particular kind of cell or tissue. Pair-rule genes are usually expressed in the primordia of alternate segments and are required for eventual formation of those same segments. paired (prd) is conserved as a pair-rule gene in phylogenetically diverse insects, such as in beetles and fruit flies, and is required for their normal segmentation and embryonic viability. When comparing segmentation mechanisms between the fruit fly, Drosophila melanogaster, and the mosquito, Anopheles stephensi, Dr. Jarvela strikingly found out that mosquitoes don’t have paired gene in their genomes. While insects typically have three pax3/7 genes, only two appeared to be in the many mosquito genomes she searched. These two genes have been identified as gooseberry (gsb) and gooseberry-neuro (gsb-n), as determined by phylogenetic analysis; neither of the pax3/7 genes found in mosquito genomes group with the paired genes (figure 1). Furthermore, gsb and gsb-n are syntenic – they occur adjacent to each other on the same chromosome – in the Anopheles genome, just as other gsb and gsb-n genes are in other insect genomes. Of course, Anopheles embryos and larvae retain a segmented body plan and segment polarity expression of engrailed despite the loss of prd. So, how do mosquitoes maintain their segmented body plan? To address this question, Dr. Jarvela sought to understand the roles of some of the other segmentation gene orthologs in A. stephensi. Might there be differences in their expression patterns that could explain mosquitoes’ loss of prd? Dr. Jarvela used a technique known as in situ hybridization staining, which allows one to visualize which cells contain mRNA of interest. Imaging revealed the expression patterns of most of the segmentation genes she examined were nearly identical to the expression patterns of these genes in Drosophila. She then wondered which transcription factor(s) might be taking the place of prd in the activation of engrailed, a segment polarity gene. Before she could try to identify the CREs dependent on Prd’s replacement in the mosquito, she first needed to map the Prd-dependent CREs in Drosophila. To do this, she conducted a classic enhancer-reporter screen. In this approach, a candidate CRE is placed upstream of some reporter gene, and this construct is then integrated into the Drosophila genome (Yeh et al. 1995). The ability of the candidate CRE to activate transcription of the reporter is then assayed by analyzing expression of the reporter (such as by in situ hybridization, described above). Using this technique, Dr. Jarvela was able to identify Prd-dependent engrailed regulatory regions. She then tested candidate Aste-engrailed CREs in Drosophila embryos, and was able to drive expression of a reporter in an engrailed-like pattern, demonstrating that these DNA sequences are very likely Aste-engrailed regulatory elements. Future work will look at which transcription factors might be responsible for binding these CREs in the absence of prd in mosquitoes. 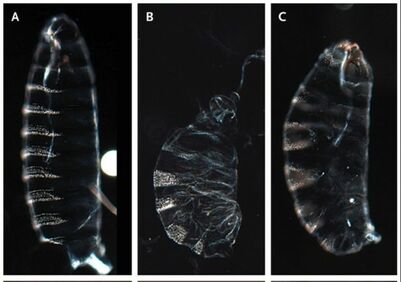 Figure 2. The pR92W NR5A1 variant is able to bind DNA and disrupt segmentation only when expressed at very high levels. A) A control embryo showing the wild-type pattern of denticle belts, B) An embryo expressing the human gene NR5A1 at six times the baseline level; this reference allele causes segmentation defects when expressed in the Drosophila embryo because it is fully able to bind Ftz-f1 binding sites but cannot activate transcription of Ftz-F1 target genes, and C) An embryo expressing the NR5A1 variant p.R92W at six times the baseline level. When expressed at three times the baseline level, the same allele does not produce segmentation defects because it does not bind the DNA well enough to out-compete the endogenous Ftz-F1, compared to the reference NR5A1 allele which produces strong segmentation defects when expressed at three times the baseline level. Figure from Splinter et al (2018). Figure 2. The pR92W NR5A1 variant is able to bind DNA and disrupt segmentation only when expressed at very high levels. A) A control embryo showing the wild-type pattern of denticle belts, B) An embryo expressing the human gene NR5A1 at six times the baseline level; this reference allele causes segmentation defects when expressed in the Drosophila embryo because it is fully able to bind Ftz-f1 binding sites but cannot activate transcription of Ftz-F1 target genes, and C) An embryo expressing the NR5A1 variant p.R92W at six times the baseline level. When expressed at three times the baseline level, the same allele does not produce segmentation defects because it does not bind the DNA well enough to out-compete the endogenous Ftz-F1, compared to the reference NR5A1 allele which produces strong segmentation defects when expressed at three times the baseline level. Figure from Splinter et al (2018). In addition to her work on mosquitoes, Dr. Jarvela also shared some of her work in Drosophila melanogaster which demonstrated the importance of the species as a model organism for understanding human genetic diseases. In a recent collaboration with the Undiagnosed Diseases Network, Dr. Jarvela and the Pick lab were contacted in an effort to use the fruit fly to study a newly identified human polymorphism in a certain transcription factor called NR5A1. In humans, this transcription factor affects the activity of several gonad development genes (Bashamboo 2016). In fruit flies, this gene is known as ftz-f1 and is a pair-rule gene, required for development of alternate segments similar to prd. The identified genetic change affects a DNA-contact point, which could either ablate or weaken DNA binding function. Dr. Jarvela could express this human NR5A1 (ftz-f1) variant allele in fruit flies and observe its effects on fruit fly embryo development. She used a driver gene system (GAL4-UAS) so she could control the level of the protein’s expression, which led her to the observation that the NR5A1 variant allele’s effects were dose-dependent - meaning high amounts of the protein were needed before segmentation defects were observed in Drosophila embryos. This is suggestive of the genetic change of the transcription factor being a partial loss of function, with some function still present. This led to a better understanding of the genetics underlying a new human syndrome (figure 2) (Bashamboo 2016, Splinter 2018). References
Comments are closed.
|
Categories
All
Archives
June 2024
|
Department of Entomology
University of Maryland
4112 Plant Sciences Building
College Park, MD 20742-4454
USA
Telephone: 301.405.3911
Fax: 301.314.9290
University of Maryland
4112 Plant Sciences Building
College Park, MD 20742-4454
USA
Telephone: 301.405.3911
Fax: 301.314.9290

 RSS Feed
RSS Feed




