|
The Greek philosopher Heraclitus was channeling his inner taxonomist when he declared, “everything is in flux.” Depending on who you ask, taxonomy as a field precariously teeters between being either the foundation of all other biological sciences or the most esoteric debate topic two scientists can choose. Scientists rely on taxonomists to draw the lines between the species that we study so we can all be on the same page while we study them (imagine trying to plan a trip to the zoo without names for the animals). As Dr. Jason Mottern explained during the last Entomology Department colloquium of 2016, these lines are often drawn in pencil.
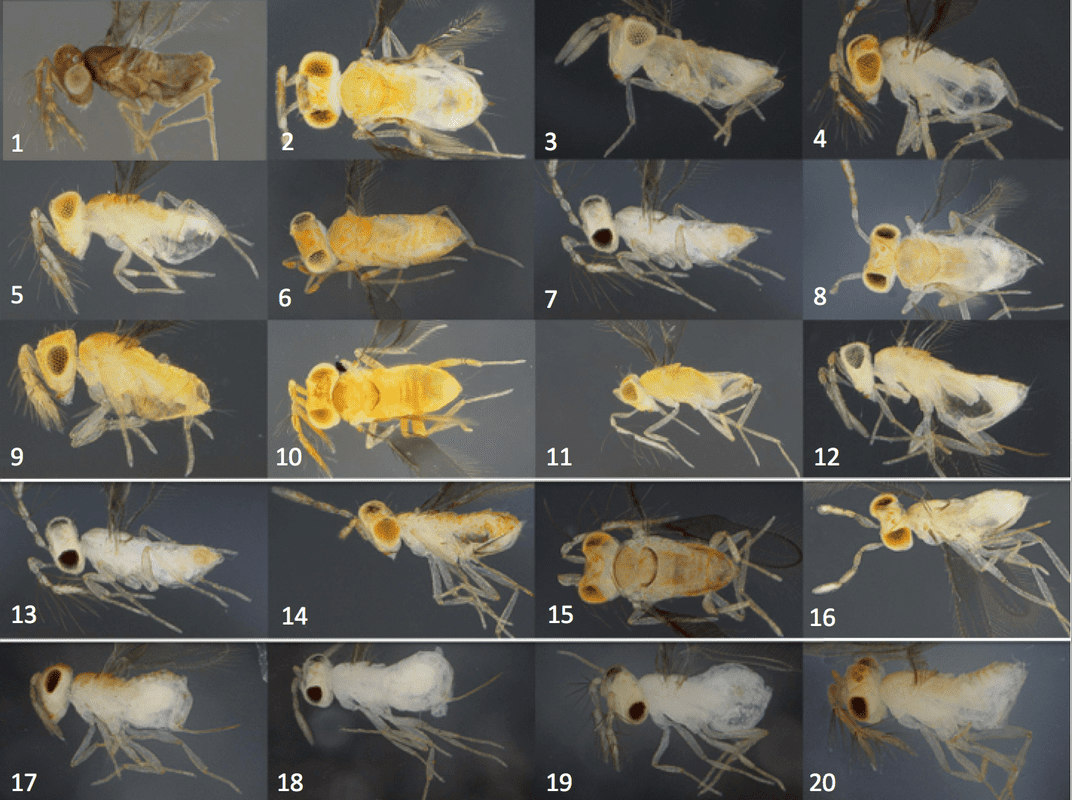
proliferation of DNA sequencing, many organisms once classified as the same primarily on similarities in observable traits like shape, size, and color are now being reconsidered. However, it can be difficult to observe these characteristics in organisms that are very small, like the wasps Dr. Mottern studies. Organisms can also evolve to look similar when they are actually not very closely related in a process called convergent evolution. The similar appearance of unrelated organisms is frequently the result of a shared lifestyle or mimicry to confuse predators, and it is just as likely to confuse taxonomists. Today, taxonomists use both physical observations and DNA sequencing to try to identify new species and correct taxonomic mistakes of the past. Dr. Mottern described one such taxonomic revision during his seminar that began as a disagreement between two researchers at University of California (UC), Riverside. Both researchers believed they had sampled the same species, Cales noacki, at the UC Riverside Biological Control Grove, yet the two sets of DNA data did not agree. While the two researchers were attacking the molecular biology skills of the other, Dr. Mottern stepped in to help prove they were both right: what if two species of wasp had been masquerading as a single species? To answer this question, Dr. Mottern needed the thing every taxonomist needs: more samples. As a result, he placed a the call for an assortment of wasp species in the genus Cales using this charming ‘wanted poster’ at an entomology meeting (Fig 1).
voucher specimens from biological control programs, to confirm that one of the “Cales noacki” shipments contained a species indistinguishable in appearance, but distinct according to DNA analysis, living cryptically among the true Cales noacki of the UC Riverside Biological Grove. Dr. Mottern named this species Cales rosei (Fig. 3), but he had no idea where this little wasp originally came from. The trouble was, there had been many subsequent shipments in the 1960s. Cales noacki had quite the knack for killing woolly whiteflies, a serious foliar pest of citrus in California. Based on quarantine and release records, the population introduced to UC Riverside is actually a mixture of specimens from Chile, Peru, Brazil and Argentina. All of these shipments aimed to establish a vibrant community of Cales noacki that could be studied and shipped around the world to control whiteflies, but one shipment must have been misidentified: which shipment brought Cales rosei to this Cales-less grove in California? To answer this question, Dr. Mottern turned to the slides prescient entomologists had kept as a record of each previous shipment: each time a new batch of wasps was incorporated, a handful of wasps were stored on a slide as a historical record.
Aside from resolving future systematic disputes, Dr. Mottern seems to have his eyes set on clarifying the so-called “phylogenetic backbone” of these small, parasitic wasps. The phylogenetic backbone refers to the greater genus and family level relatedness. He hopes that by incorporating even more information, through next-generation sequencing, taxonomists can resolve the relationships between each group of these wasps - something about which scientists still currently know very little.
References Mottern, J. L., & Heraty, J. M. (2014). The dead can talk: Museum specimens show the origins of a cryptic species used in biological control. Biological Control, 71, 30-39. Heraty, J. M., Burks, R. A., Cruaud, A., Gibson, G. A., Liljeblad, J., Munro, J., ... & Huber, J. (2013). A phylogenetic analysis of the megadiverse Chalcidoidea (Hymenoptera). Cladistics, 29(5), 466-542. Heraty, J., Ronquist, F., Carpenter, J. M., Hawks, D., Schulmeister, S., Dowling, A. P., ... & Sharkey, M. (2011). Evolution of the hymenopteran megaradiation. Molecular phylogenetics and evolution, 60(1), 73-88. Rosen, D., & DeBach, P. (1976). Biosystematic studies on the species of Aphytis (Hymenoptera: Aphelinidae). Mushi, 49(1), 1-17. Zelditch, M. L., Swiderski, D. L., & Sheets, H. D. (2012). Geometric morphometrics for biologists: a primer. Academic Press. About the authors: Brian Lovett is a PhD student in Dr. Raymond St. Leger’s Lab studying mycology and genetics in agricultural and vector biology systems. He is currently working on projects analyzing mycorrhizal interactions in agricultural systems, the transcriptomics of malaria vector mosquitoes, and the genomes of entomopathogenic fungi. Mengyao Chen is a Master’s student in Dr. Leslie Pick’s Lab. Her research focuses on segmentation genes in Brown Marmorated Stink Bug (BMSB, Halyomorpha halys). Her current work is looking for orthologs of pair-rule genes in BMSB, and studying their expression and functions using in situ hybridization and RNAi. Comments are closed.
|
Categories
All
Archives
June 2024
|
Department of Entomology
University of Maryland
4112 Plant Sciences Building
College Park, MD 20742-4454
USA
Telephone: 301.405.3911
Fax: 301.314.9290
University of Maryland
4112 Plant Sciences Building
College Park, MD 20742-4454
USA
Telephone: 301.405.3911
Fax: 301.314.9290




 RSS Feed
RSS Feed




